AlphaFold 3: Revolutionizing Molecular Structure and Interaction Prediction
Since its launch in 2020, over 2 million researchers have turned to Google DeepMind's AlphaFold 2 to predict protein structures, aiding in breakthroughs like vaccine development and cancer treatments. This model tackled a challenge that had stumped scientists for over half a century. But the team at Google DeepMind didn't stop there; instead of resting on their laurels, they rolled up their sleeves and started working on AlphaFold 3.
Launched in May by Google DeepMind and Isomorphic Labs, AlphaFold 3 takes things up a notch. It doesn't just predict protein folding; it also forecasts the structure and interactions of all sorts of life's molecules, including DNA, RNA, and ligands—those tiny molecules that attach to proteins.
"With AlphaFold 2, we made huge strides in solving the protein folding puzzle, but the scientific community has moved on to more complex stuff," explains Jonas Adler, a research scientist at Google DeepMind. "Researchers are now diving into details like how small molecules bind, or how RNA works, areas where AlphaFold 2 fell short. To keep up with the latest in biology and chemistry, we needed a tool that could handle all kinds of biomolecules."
"Everything" here includes ligands, which are crucial because they make up about half of all drugs. "At Isomorphic Labs, we're seeing the huge potential of AlphaFold 3 for designing drugs rationally, and we're already putting it to use every day," says Adrian Stecula, a research leader at Isomorphic Labs. "We're exploring how new small molecules bind to new drug targets, figuring out how proteins interact with DNA and RNA, and studying how chemical tweaks affect protein structures—the new model opens up all these possibilities."
Incorporating these extra molecule types meant dealing with a ton more combinations. "Proteins are pretty straightforward; there are only 20 standard amino acids," Jonas notes. "But small molecules? They're all over the place, with endless possibilities. They're super diverse."
Realizing that a comprehensive database was out of the question, the team launched AlphaFold Server. This free tool lets scientists input their own sequences, and AlphaFold generates the molecular complexes for them. Since going live in May, it's been used to create over 1 million structures.
"It's like Google Maps but for molecular complexes," says Lindsay Willmore, a research engineer at Google DeepMind. "Even if you're not a coder, you can just copy and paste your protein, DNA, RNA, or small molecule sequences, hit a button, and wait a few minutes. You'll get your structure along with confidence metrics to help you evaluate the prediction."
To make AlphaFold 3 work with such a broad range of biomolecules, the team expanded the training data to include DNA, RNA, small molecules, and more. "We figured, 'Why not train on everything in our dataset that helped us with proteins and see how far we can go?'" Lindsay explains. "Turns out, we went pretty far."
A key change in AlphaFold 3 was in the final part of the model that creates the structure. While AlphaFold 2 used a complex custom geometry-based module, AlphaFold 3 switched to a simpler generative model based on diffusion, similar to other advanced image generation models like Imagen. This change streamlined how the model deals with the new types of molecules.
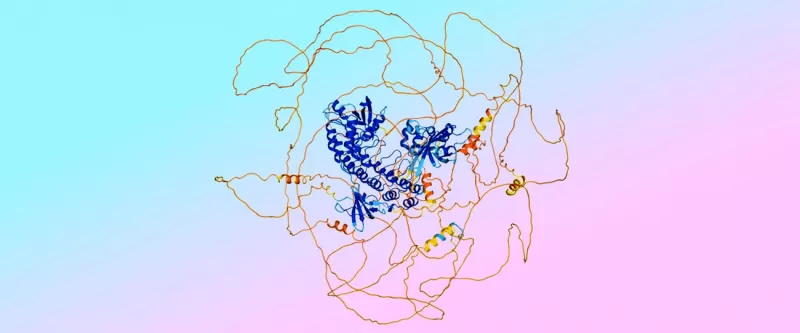
However, this shift brought a new challenge: the diffusion model would mistakenly create an "ordered" structure for protein regions that are actually "disordered"—think of it as trying to organize a pile of chaotic spaghetti into a neat spiral.
So, the team turned to AlphaFold 2, which excels at predicting these disordered interactions. "We used those predictions from AlphaFold 2 to train AlphaFold 3, teaching it to recognize and predict disorder," Lindsay says.
"We have a saying: 'Trust the fusilli, reject the spaghetti,'" adds Jonas with a chuckle.
An example of a prediction from AlphaFold 3, showing ordered “fusilli” regions in blue and disordered “spaghetti” regions in orange. The colors indicate the model's confidence in the prediction's accuracy.
The team is excited to see how AlphaFold 3 will be used in fields like genomics and drug design. "It's amazing to see how far we've come," Jonas says. "What was once tough is now easy, and what was impossible is now within reach. There are still tough nuts to crack, but we're thrilled about what AlphaFold 3 can help us achieve."
Related article
 "Dot AI Companion App Announces Closure, Discontinues Personalized Service"
Dot, an AI companion application designed to function as a personal friend and confidant, will cease operations, according to a Friday announcement from its developers. New Computer, the startup behind Dot, stated on its website that the service will
"Dot AI Companion App Announces Closure, Discontinues Personalized Service"
Dot, an AI companion application designed to function as a personal friend and confidant, will cease operations, according to a Friday announcement from its developers. New Computer, the startup behind Dot, stated on its website that the service will
 Anthropic Resolves Legal Case Over AI-Generated Book Piracy
Anthropic has reached a resolution in a significant copyright dispute with US authors, agreeing to a proposed class action settlement that avoids a potentially costly trial. The agreement, filed in court documents this Tuesday, stems from allegations
Anthropic Resolves Legal Case Over AI-Generated Book Piracy
Anthropic has reached a resolution in a significant copyright dispute with US authors, agreeing to a proposed class action settlement that avoids a potentially costly trial. The agreement, filed in court documents this Tuesday, stems from allegations
 Figma Releases AI-Powered App Builder Tool to All Users
Figma Make, the innovative prompt-to-app development platform unveiled earlier this year, has officially exited beta and rolled out to all users. This groundbreaking tool joins the ranks of AI-powered coding assistants like Google's Gemini Code Assis
Comments (21)
0/200
Figma Releases AI-Powered App Builder Tool to All Users
Figma Make, the innovative prompt-to-app development platform unveiled earlier this year, has officially exited beta and rolled out to all users. This groundbreaking tool joins the ranks of AI-powered coding assistants like Google's Gemini Code Assis
Comments (21)
0/200
![HarryClark]() HarryClark
HarryClark
 August 21, 2025 at 3:01:19 AM EDT
August 21, 2025 at 3:01:19 AM EDT
AlphaFold 3 sounds like a game-changer! Predicting molecular interactions could speed up drug discovery big time. Curious how it stacks up against traditional lab methods. 🧬


 0
0
![PeterThomas]() PeterThomas
PeterThomas
 August 16, 2025 at 7:01:00 PM EDT
August 16, 2025 at 7:01:00 PM EDT
AlphaFold 3 sounds like a game-changer! Predicting molecular interactions with such precision could really speed up drug discovery. I'm curious how it stacks up against traditional lab methods in terms of cost and time. Anyone know? 🤔


 0
0
![JoseLewis]() JoseLewis
JoseLewis
 August 14, 2025 at 7:00:59 AM EDT
August 14, 2025 at 7:00:59 AM EDT
AlphaFold 3 sounds like a game-changer! Predicting molecular interactions could speed up drug discovery big time. Excited to see how it shapes medical breakthroughs! 🚀


 0
0
![ElijahCollins]() ElijahCollins
ElijahCollins
 August 13, 2025 at 11:01:00 PM EDT
August 13, 2025 at 11:01:00 PM EDT
AlphaFold 3 sounds like a game-changer! Predicting molecular interactions could speed up drug discovery big time. Curious how it stacks against human researchers—will it outsmart us or just make our coffee breaks longer? 😄


 0
0
![GregorySmith]() GregorySmith
GregorySmith
 August 5, 2025 at 10:01:00 PM EDT
August 5, 2025 at 10:01:00 PM EDT
AlphaFold 3 sounds like a game-changer! Predicting molecular interactions with such precision could unlock new frontiers in medicine. But I wonder, will this tech stay accessible to all researchers, or get locked behind paywalls? 🤔


 0
0
![BruceThomas]() BruceThomas
BruceThomas
 August 1, 2025 at 2:08:50 AM EDT
August 1, 2025 at 2:08:50 AM EDT
AlphaFold 3 sounds like a game-changer for science! Predicting molecular interactions could speed up drug discovery big time. Excited to see where this goes! 🚀


 0
0

Since its launch in 2020, over 2 million researchers have turned to Google DeepMind's AlphaFold 2 to predict protein structures, aiding in breakthroughs like vaccine development and cancer treatments. This model tackled a challenge that had stumped scientists for over half a century. But the team at Google DeepMind didn't stop there; instead of resting on their laurels, they rolled up their sleeves and started working on AlphaFold 3.
Launched in May by Google DeepMind and Isomorphic Labs, AlphaFold 3 takes things up a notch. It doesn't just predict protein folding; it also forecasts the structure and interactions of all sorts of life's molecules, including DNA, RNA, and ligands—those tiny molecules that attach to proteins.
"With AlphaFold 2, we made huge strides in solving the protein folding puzzle, but the scientific community has moved on to more complex stuff," explains Jonas Adler, a research scientist at Google DeepMind. "Researchers are now diving into details like how small molecules bind, or how RNA works, areas where AlphaFold 2 fell short. To keep up with the latest in biology and chemistry, we needed a tool that could handle all kinds of biomolecules."
"Everything" here includes ligands, which are crucial because they make up about half of all drugs. "At Isomorphic Labs, we're seeing the huge potential of AlphaFold 3 for designing drugs rationally, and we're already putting it to use every day," says Adrian Stecula, a research leader at Isomorphic Labs. "We're exploring how new small molecules bind to new drug targets, figuring out how proteins interact with DNA and RNA, and studying how chemical tweaks affect protein structures—the new model opens up all these possibilities."
Incorporating these extra molecule types meant dealing with a ton more combinations. "Proteins are pretty straightforward; there are only 20 standard amino acids," Jonas notes. "But small molecules? They're all over the place, with endless possibilities. They're super diverse."
"It's like Google Maps but for molecular complexes," says Lindsay Willmore, a research engineer at Google DeepMind. "Even if you're not a coder, you can just copy and paste your protein, DNA, RNA, or small molecule sequences, hit a button, and wait a few minutes. You'll get your structure along with confidence metrics to help you evaluate the prediction."
To make AlphaFold 3 work with such a broad range of biomolecules, the team expanded the training data to include DNA, RNA, small molecules, and more. "We figured, 'Why not train on everything in our dataset that helped us with proteins and see how far we can go?'" Lindsay explains. "Turns out, we went pretty far."
A key change in AlphaFold 3 was in the final part of the model that creates the structure. While AlphaFold 2 used a complex custom geometry-based module, AlphaFold 3 switched to a simpler generative model based on diffusion, similar to other advanced image generation models like Imagen. This change streamlined how the model deals with the new types of molecules.
However, this shift brought a new challenge: the diffusion model would mistakenly create an "ordered" structure for protein regions that are actually "disordered"—think of it as trying to organize a pile of chaotic spaghetti into a neat spiral.
So, the team turned to AlphaFold 2, which excels at predicting these disordered interactions. "We used those predictions from AlphaFold 2 to train AlphaFold 3, teaching it to recognize and predict disorder," Lindsay says.
"We have a saying: 'Trust the fusilli, reject the spaghetti,'" adds Jonas with a chuckle.
The team is excited to see how AlphaFold 3 will be used in fields like genomics and drug design. "It's amazing to see how far we've come," Jonas says. "What was once tough is now easy, and what was impossible is now within reach. There are still tough nuts to crack, but we're thrilled about what AlphaFold 3 can help us achieve."
 Anthropic Resolves Legal Case Over AI-Generated Book Piracy
Anthropic has reached a resolution in a significant copyright dispute with US authors, agreeing to a proposed class action settlement that avoids a potentially costly trial. The agreement, filed in court documents this Tuesday, stems from allegations
Anthropic Resolves Legal Case Over AI-Generated Book Piracy
Anthropic has reached a resolution in a significant copyright dispute with US authors, agreeing to a proposed class action settlement that avoids a potentially costly trial. The agreement, filed in court documents this Tuesday, stems from allegations
 Figma Releases AI-Powered App Builder Tool to All Users
Figma Make, the innovative prompt-to-app development platform unveiled earlier this year, has officially exited beta and rolled out to all users. This groundbreaking tool joins the ranks of AI-powered coding assistants like Google's Gemini Code Assis
Figma Releases AI-Powered App Builder Tool to All Users
Figma Make, the innovative prompt-to-app development platform unveiled earlier this year, has officially exited beta and rolled out to all users. This groundbreaking tool joins the ranks of AI-powered coding assistants like Google's Gemini Code Assis
 August 21, 2025 at 3:01:19 AM EDT
August 21, 2025 at 3:01:19 AM EDT
AlphaFold 3 sounds like a game-changer! Predicting molecular interactions could speed up drug discovery big time. Curious how it stacks up against traditional lab methods. 🧬


 0
0
 August 16, 2025 at 7:01:00 PM EDT
August 16, 2025 at 7:01:00 PM EDT
AlphaFold 3 sounds like a game-changer! Predicting molecular interactions with such precision could really speed up drug discovery. I'm curious how it stacks up against traditional lab methods in terms of cost and time. Anyone know? 🤔


 0
0
 August 14, 2025 at 7:00:59 AM EDT
August 14, 2025 at 7:00:59 AM EDT
AlphaFold 3 sounds like a game-changer! Predicting molecular interactions could speed up drug discovery big time. Excited to see how it shapes medical breakthroughs! 🚀


 0
0
 August 13, 2025 at 11:01:00 PM EDT
August 13, 2025 at 11:01:00 PM EDT
AlphaFold 3 sounds like a game-changer! Predicting molecular interactions could speed up drug discovery big time. Curious how it stacks against human researchers—will it outsmart us or just make our coffee breaks longer? 😄


 0
0
 August 5, 2025 at 10:01:00 PM EDT
August 5, 2025 at 10:01:00 PM EDT
AlphaFold 3 sounds like a game-changer! Predicting molecular interactions with such precision could unlock new frontiers in medicine. But I wonder, will this tech stay accessible to all researchers, or get locked behind paywalls? 🤔


 0
0
 August 1, 2025 at 2:08:50 AM EDT
August 1, 2025 at 2:08:50 AM EDT
AlphaFold 3 sounds like a game-changer for science! Predicting molecular interactions could speed up drug discovery big time. Excited to see where this goes! 🚀


 0
0